Setup
A log file is created in the DIAGNOSTIC_FOLDER, where all pipeline steps will be logged, along with raw prompt requests and responses.The pipeline begins by looking for PDFs in the data directory.PDF’s are validated to ensure metadata access.
Preprocess pages

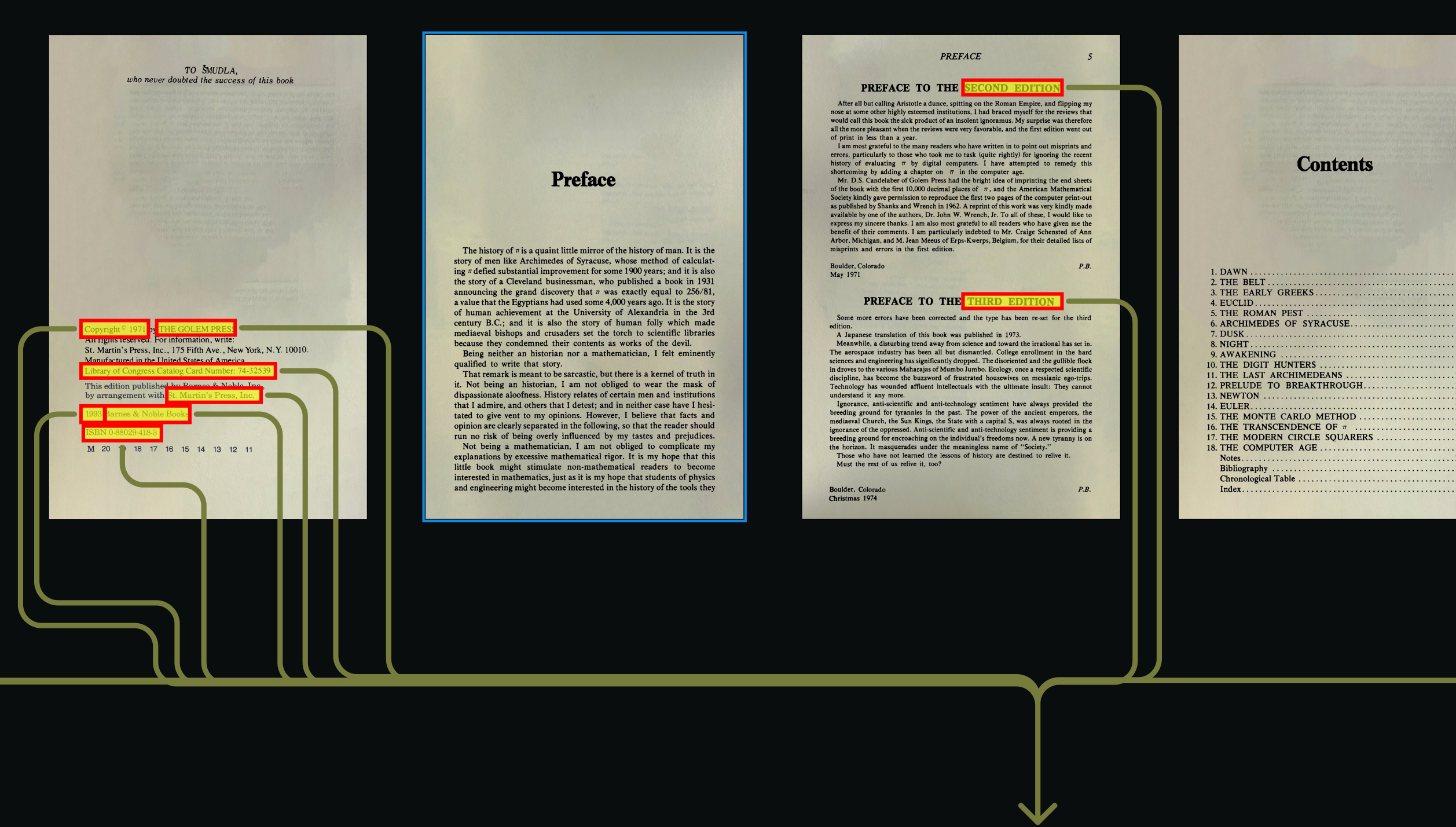
PyMuPDF Python library to identify the front and back matter pages. What are matter pages?By default, the first 8 front matter pages are processed. Body matter is always ignored, and back matter inclusion can be optionally configured with MATTER_CONFIG.back settings.Processed page images are then enhanced for OCR with the PIL (Pillow) Python library.Original and enhanced images are written to the diagnostic folder when SAVE_DIAG_PER_PAGE_IMG=true.Run traditional OCR

text, image, or mixed). Tesseract OCR runs and quality is scored with OCR_CONFIDENCE_THRESHOLD values.This produces raw text content but doesn’t preserve document structure or hierarchy.The extracted, unstructured text files are written to the diagnostic folder
when SAVE_DIAG_TXT_PER_PG=true.Extract structured text
Analyze with a VLM
When ANALYZE_WITH_VISION=true,
the page is analyzed by a Vision Language Model that holistically
understands the page elements and their relationships.We use Meta’s
llama3.2-vision, hosted by Ollama locally, on your network or in the cloud.
Setup and configuration instructions.Analysis text files are written to the diagnostic folder when SAVE_DIAG_TXT_PER_PG=true.
This step is in active development, and VLM results are not yet integrated into the pipeline.
Compose metadata prompt
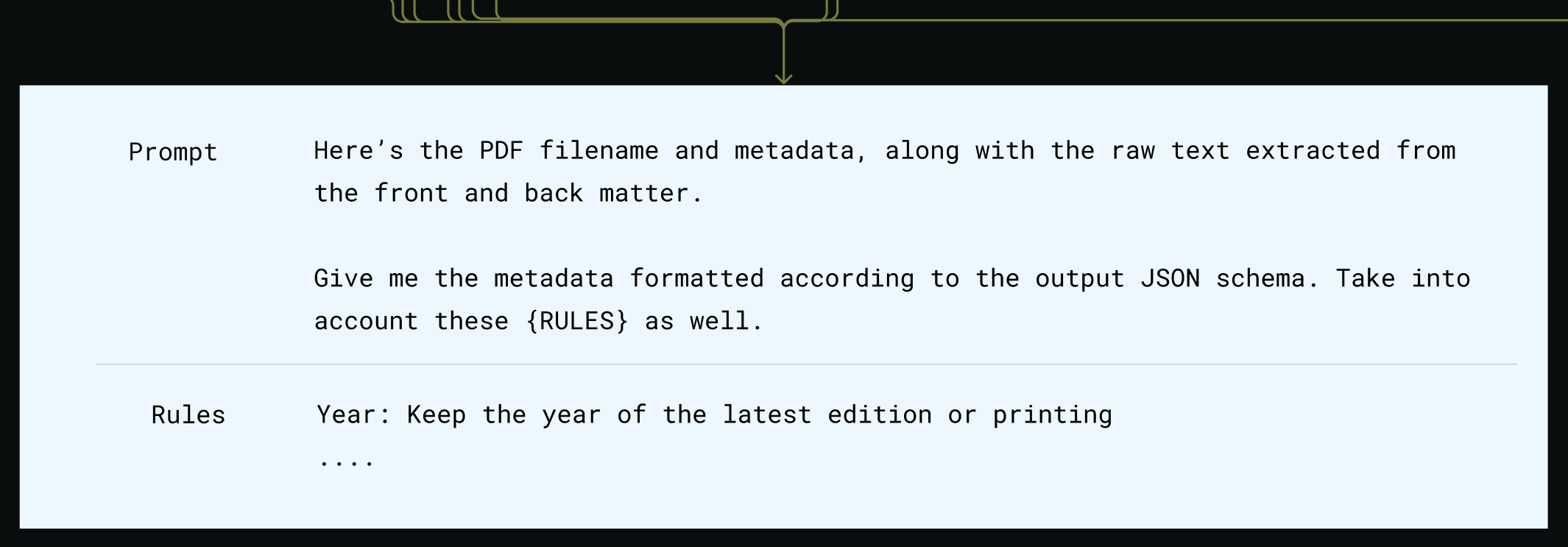
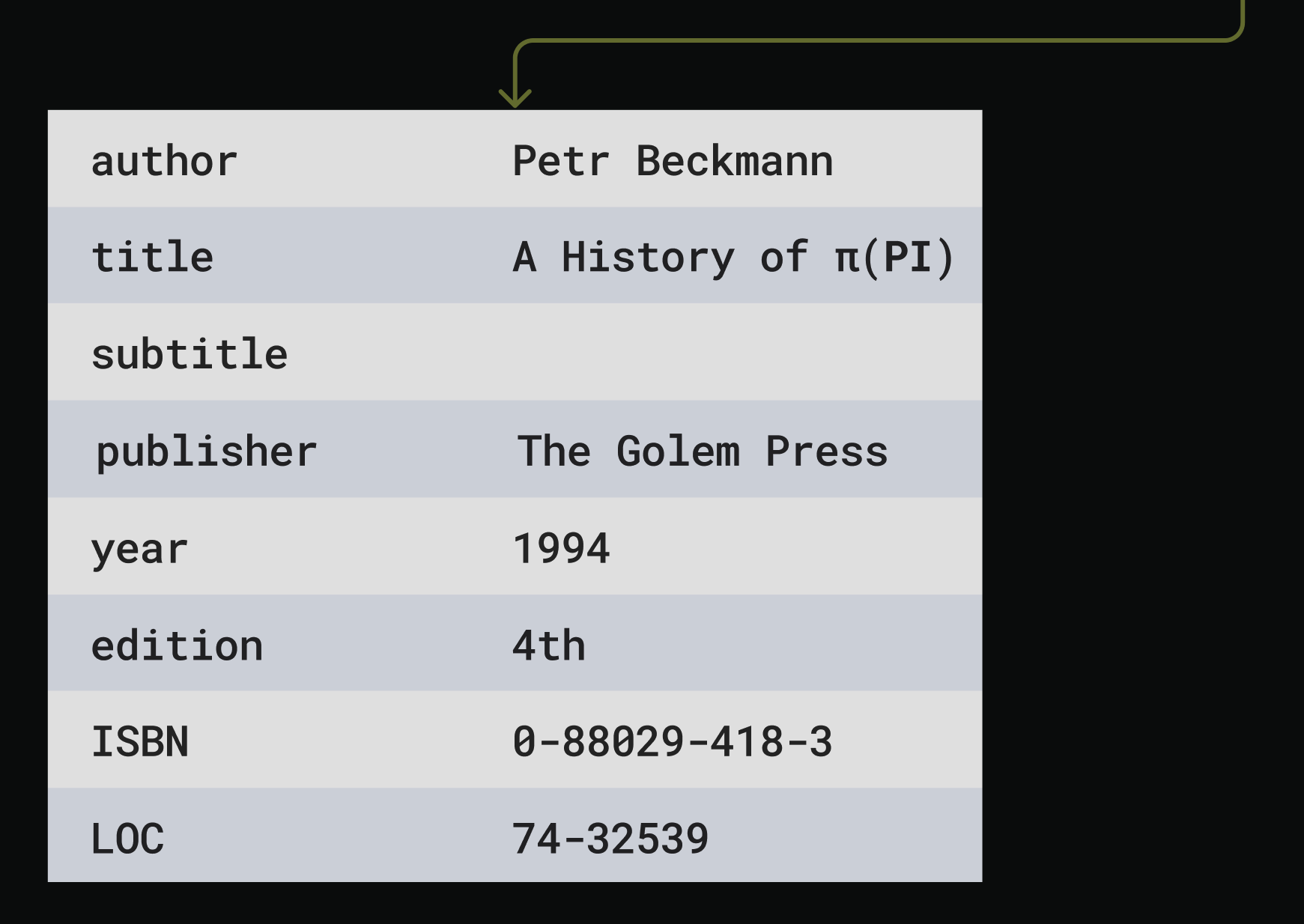
SELECTED_MODEL_NAME.The prompt includes detailed extraction rules, ie, what to do when multiple editions are found.Process metadata response
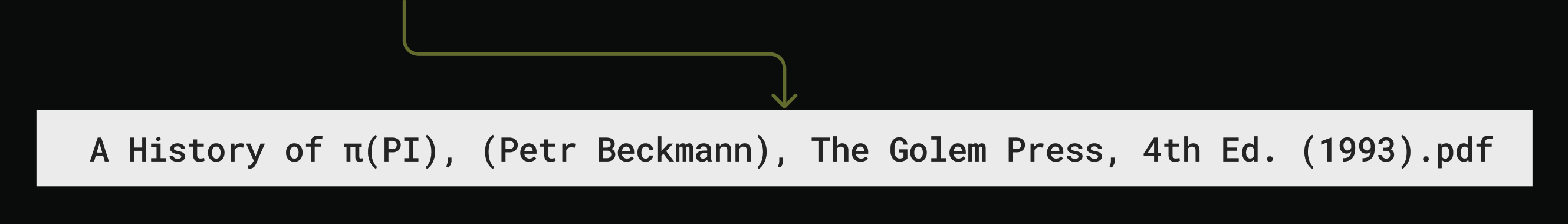
Compose filename prompt
The prompt for filename generation is sent to OpenAI,
with detailed format constraints:
Process filename response
Annotation Export
When we pass--with-annot-export to the main CLI the processor writes UTF-8 sidecar files beside every annotated PDF.
The exporter names the markdown file after the cleaned destination filename, swaps .pdf for --ann.md, and overwrites
on every run so repeated passes stay deterministic. Sidecars live next to the PDFs so downstream sync tooling can
collect them without new path logic. We also generate <filename>--ann.json, which captures the geometry, colors,
and metadata needed to reconstruct the annotations later.
Sometimes we want annotation text without running the full metadata flow. Run the standalone helper below to walk a
directory and create sidecars that mirror the original filenames. The command accepts a single PDF path or a
directory of PDFs, matching file names case-insensitively.
extract_annotations_complex so they capture highlights, strikeouts, and freeform notes exactly as
PyMuPDF exposes them. The JSON sidecar keeps placement and styling data so we can replay annotations onto a matching PDF.